Scientific research has reached unprecedented new parallels due to the development of the computer. One field that has seen significant advancement is genetics. Bioinformatics uses computational tools to better understand genetic data. The most famous bioinformatics project is the human genome project.
“[The Human Genome Project was] launched in October 1990 and completed in April 2003…”
The Human Genome Project
The basis of the human genome project was to sequence complete human DNA. This has laid a base for advancements in genetics, medicine, and other scientific fields.
String Matching
The basis of bioinformatics is matching a sequence of genes. Although this sounds difficult, it has been a common study for longer than computer science has been around.
Matching a sequence of characters in an alphabet is also the basis of compilers, search engines, comparators, and many other tools. Although it seems simple enough, genomes are billions or trillions of characters long. Searching for subsequences in so much text requires some clever approaches. Enter, the tree.
The Tree
Comparing all characters to find a pattern would be painfully slow. Parsing with a tree is a much better approach. However, gene sequences can reach great lengths. Storing all characters in memory is equally expensive. Bioinformatics tools must weigh the costs of speed and available resources to produce a result that is acceptable to the researcher.
Now try it!
BLAST is a freely available web application provided by the National Library of Medicine. We will use it to compare two gene sequences.
“BLAST finds regions of similarity between biological sequences.”
BLAST
We will compare two gene sequences using BLASTp (Protein BLAST). This query will return list of sequences with strong alignment, allowing the most probable organism to be determined.
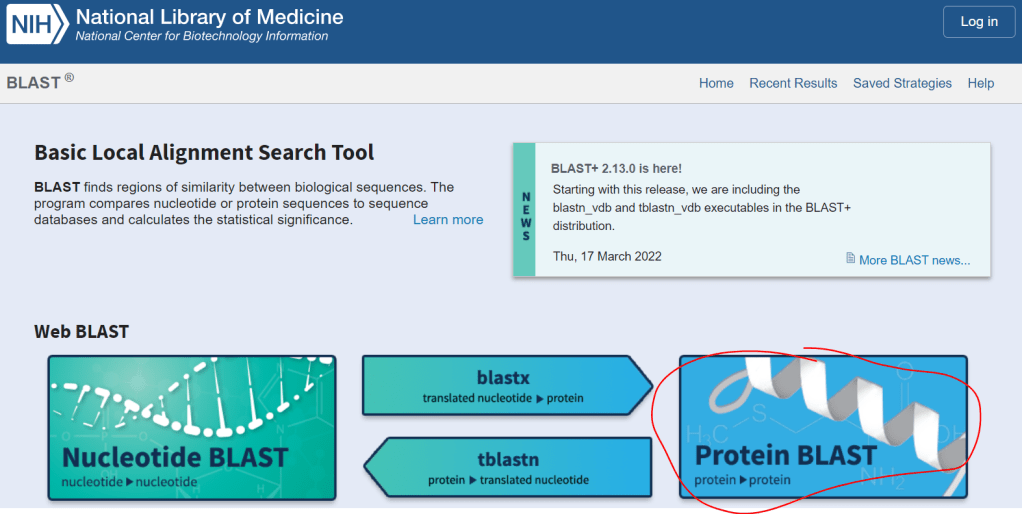
Head over to BLAST and select “Protein BLAST”.
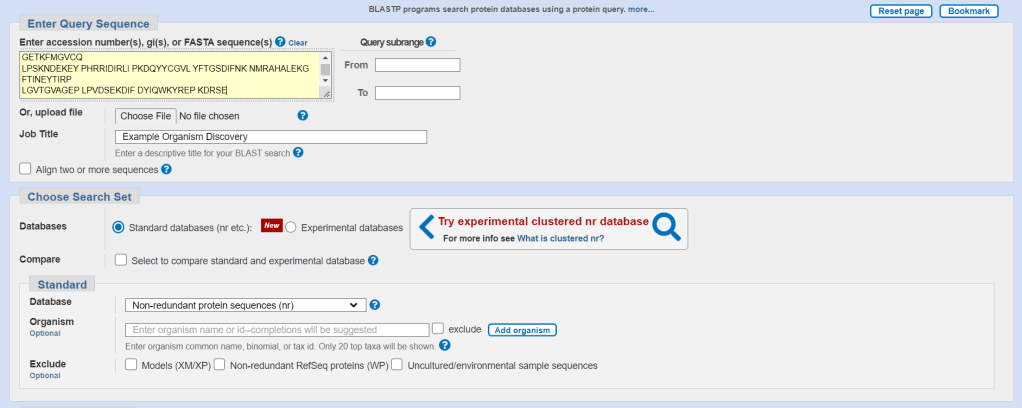
We will search for the following sequence to determine the most probable organism:
MSKRKAPQET LNGGITDMLT ELANFEKNVS QAIHKYNAYR KAASVIAKYP HKIKSGAEAK
KLPGVGTKIA EKIDEFLATG KLRKLEKIRQ DDTSSSINFL TRVSGIGPSA ARKFVDEGIK
TLEDLRKNED KLNHHQRIGL KYFGDFEKRI PREEMLQMQD IVLNEVKKVD SEYIATVCGS
FRRGAESSGD MDVLLTHPSF TSESTKQPKL LHQVVEQLQK VHFITDTLSK GETKFMGVCQ
LPSKNDEKEY PHRRIDIRLI PKDQYYCGVL YFTGSDIFNK NMRAHALEKG FTINEYTIRP
LGVTGVAGEP LPVDSEKDIF DYIQWKYREP KDRSE
The first sequence returned is always a synthetic organism and does not indicate a match.
After some time the sequences with the strongest alignment are returned.
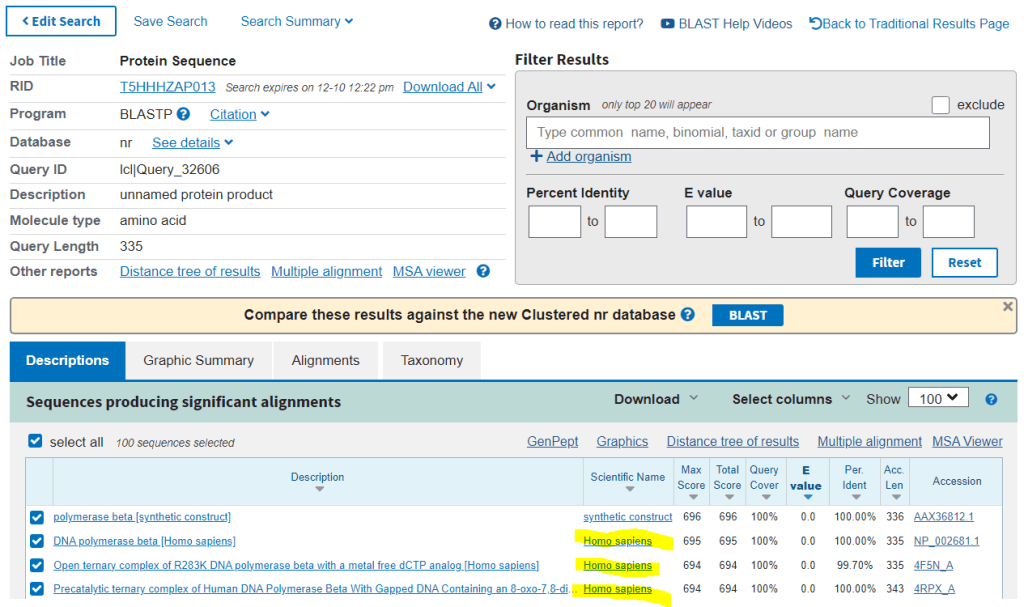
The results show that the gene sequences with the strongest alignment match homo sapiens (humans).
Real-World Application
Although the example above demonstrates matching a sequence of genes, identifying the donor organism is seldom the objective. Instead, the most common application of bioinformatics is identifying germline mutations in humans.
“Mutations in these cells are the only mutations that can be passed to offspring…”
Germline Mutation
Germline mutations are the inheritable mutations that are passed from parents to children. Predicting harmful mutations is the predominant measure of this scientific endeavor. Bioinformatics allows an unprecedented prediction of disease and other negative factors. With careful application, this will allow the mitigation of many genetic diseases.
Leave a comment